The objective of the American Heart Association COVID-19 Data Challenge is to examine the relationships between COVID-19, other health conditions, health disparities and/or social determinants of health.
Stage 1:
Starts: May 12, 2020
Ends: July 12, 2020
Stage 2:
Starts: September 1, 2020
Ends: November 1, 2020
The American Heart Association ("The Association") Institute for Precision Cardiovascular Medicine seeks to better understand the relationships between COVID-19, other health conditions, health disparities, and social determinants of health. Health disparities include poverty, environmental threats, inadequate access to health care, individual and behavioral factors and educational inequalities. Social determinants of health include resources such as food supply, housing, economic and social relationships, education, and health care.
The COVID-19 Data Challenge asks participants to use the Precision Medicine Platform (PMP) to test the relationships between COVID-19, other health conditions, health disparities and social determinants of health using their own data and/or data available on the Precision Medicine Platform.
The American Heart Association COVID-19 Data Challenge includes two stages. Stage 1 will take place from May 12 - July 12, 2020. The top six applicants based on peer review will each receive $5,000. These six applicants will advance to the second stage of the American Heart Association COVID-19 Data Challenge that will take place from September 1 - November 1, 2020. The top applicant based on peer review of the second stage of the American Heart Association COVID-19 Data Challenge will receive $15,000.
Challenges will be reviewed by a peer review committee that will evaluate:
Upon registration for the data challenge, each participant/team will be provided a PMP workspace, free of charge to complete analyses for the challenge. Results must be submitted prior to 5 p.m. Central on July 12, 2020 to be considered for the data challenge. Details on submission requirements and how to submit results are provided in the submission section. Information about PMP workspaces is provided in the Getting Started section.
Follow the steps below to get started and participate in the challenge:
1. Sign up for a PMP Account
2. Challenge Registration
Note: The Workspace request portion of the challenge registration uses the standard workspace request form. As a result, some parts do not apply to participants. Workspaces are available to COVID-19 Challenge participants free of charge from May 12 - July 12, 2020
CHEERS! You are now registered on the PMP and the Data Challenge!
Participants may bring their own de-identified data sets to the PMP. Information on how to transfer data to the platform is provided in the Upload, Access, Download Data section under Getting Started in the PMP.
As, explained in the Getting Started in the PMP section, publicly available data can be accessed a variety of ways in the PMP including downloading data directly and pulling/cloning data from repositories. Additionally, Burst IQ Research Foundry, has collected a variety of datasets, including many COVID-19 datasets and made them accessible in the PMP. A table of public datasets accessible in the PMP through BurstIQ's Research Foundry is listed here. Details on how to access datasets using BurstIQ are provided in the Getting Started in the PMP section.
The Precision Medicine Platform (PMP) provides a friendly web UI that allows users to access a secure, cloud-based environment and contains a variety of standard software and packages such as Python R . The platform workspace leverages Jupyter and RStudio to allow users to create notebooks to document and display results. Each participant must submit a notebook from their individual workspace that provides the following:
More information on Jupyter Notebooks can be found here. More in formation on R Notebooks can be found here.
Submitting a Notebook
Once you have completed your analysis and created a notebook meeting the criteria outlined above follow the steps below to submit your notebook for review:
Additional Information
Note: If you haven't done so already, please register for an account on the Precision Medicine Platform (PMP)
Overview
The PMP is a secure, HIPPA compliant and FedRAMP certified cloud-based ecosystem for facilitating data sharing, collaboration, and power computing. The PMP workspace is an interactive, AWS cloud environment comprised of common tools and software used in biomedical analyses that enables users to easily store data, collaborate, and perform analyses while also having access to elastic, power compute resources on demand.
Collaboration
The platform facilitates collaboration and reproducibility by enabling users to share workspaces and conduct analyses in a private, secure cloud environment. Users can also collaborate through traditional development methods such as github.
Collaboration is made easy with the PMP because all data and analyses reside in a secure workspace for which only the participant/team representative has access, unless the participant/team representative chooses to collaborate with colleagues and share the workspace on the PMP workspace portal.
Note: It is the participant's responsibility to ensure collaborators have the appropriate data access approvals when sharing.
Power Compute In the PMP
There are multiple ways to take advantage of power compute in the Precision Medicine Platform, namely:
Default workspace architectures are designed to fit the needs of most users, however architecture sizes can be increased and customized as needed. To increase compute resources in a workspace or get assistance with power computing on the PMP, please contact us
Terms of Use
Any inventions, intellectual property, or patents resulting from this funding are governed by the AHA Patent, Intellectual Property and Technology Transfer Policy
Please contact us if at any point you are having trouble uploading or accessing data.
Upload Data to PMP
File Upload
Files can be uploaded to the platform on the workspace portal page, by clicking the upload icon. Once the files are uploaded and you have launched your workspace, the files will be found in the folder labeled scratchbucket on the Jupyter home page (path: /mnt/workspace/scratchbucket/).
SFTP Transfer
Should you have very large data files and need an SFTP set up to upload the data, please submit a request through Technical Support.
Access Data in PMP
As mentioned, public data can be accessed in a variety of ways in a PMP workspace including downloading data directly and pulling/cloning data from repositories. Steps for accessing data from download links, repositories, and BurstIQ are described below.
Downloading Data Directly in the Precision Medicine Platform
Data can be downloaded in the workspace in the same fashion as your personal computer. Add a new tab in Firefox, navigate to the download link, and click download.
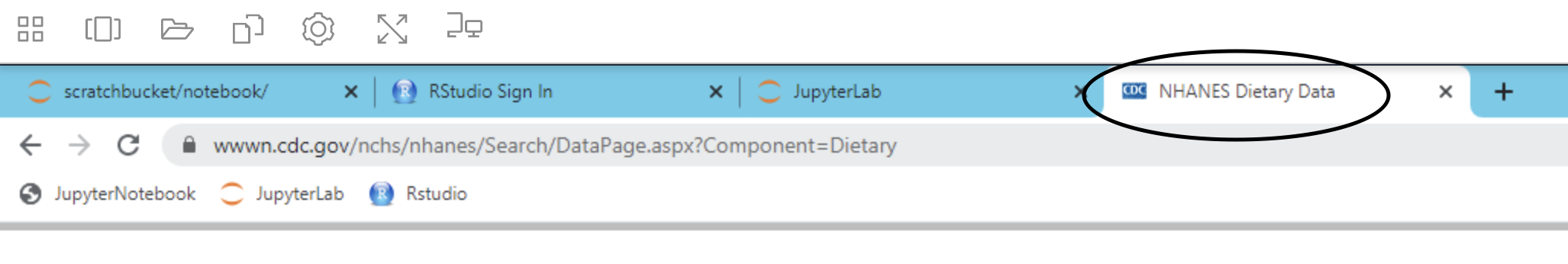
Pulling or Cloning Data from a repository
Using git repositories in the workspace functions the same way as it does on a personal computer. To pull data or clone a repository, simply go to the repository in the workspace (first tab on right shown below).
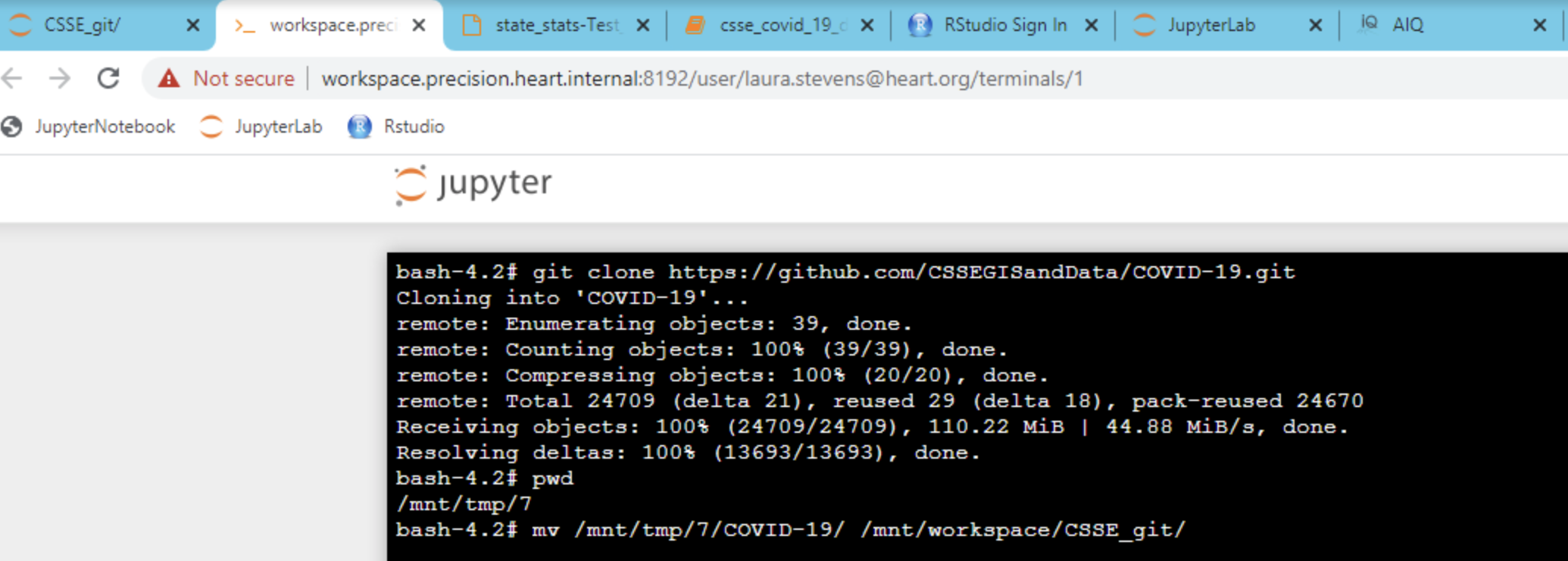
Tip: Make sure to access the repository in your workspace when setting up ssh keys. You cannot copy an ssh key out of the workspace
Accessing Data through BurstIQ
Research Foundry, hosted by BurstIQ, is a coalition of health-focused researchers, organizations, and innovators from all over the world. BurstIQ currently hosts over 100 datasets (of which 13 are COVID-19) through block chain connections. As part of the COVID-19 Data Challenge, you can request access to Research Foundry and choose any number of datasets. Information on how to register with Research Foundry and access a list of publicly available data is detailed below:
Get data out of the PMP
Data and Code within a workspace can be synced with the workspace portal by moving it to the /mnt/workspace/My_Notebooks directory within the workspace. Files located in this directory will display under the My Notebooks and Data section of workspace portal.
Note: Due to the secure nature of the workspace, certain files will not sync with the workspace portal. To extract this data, please file a ticket with Technical Support.
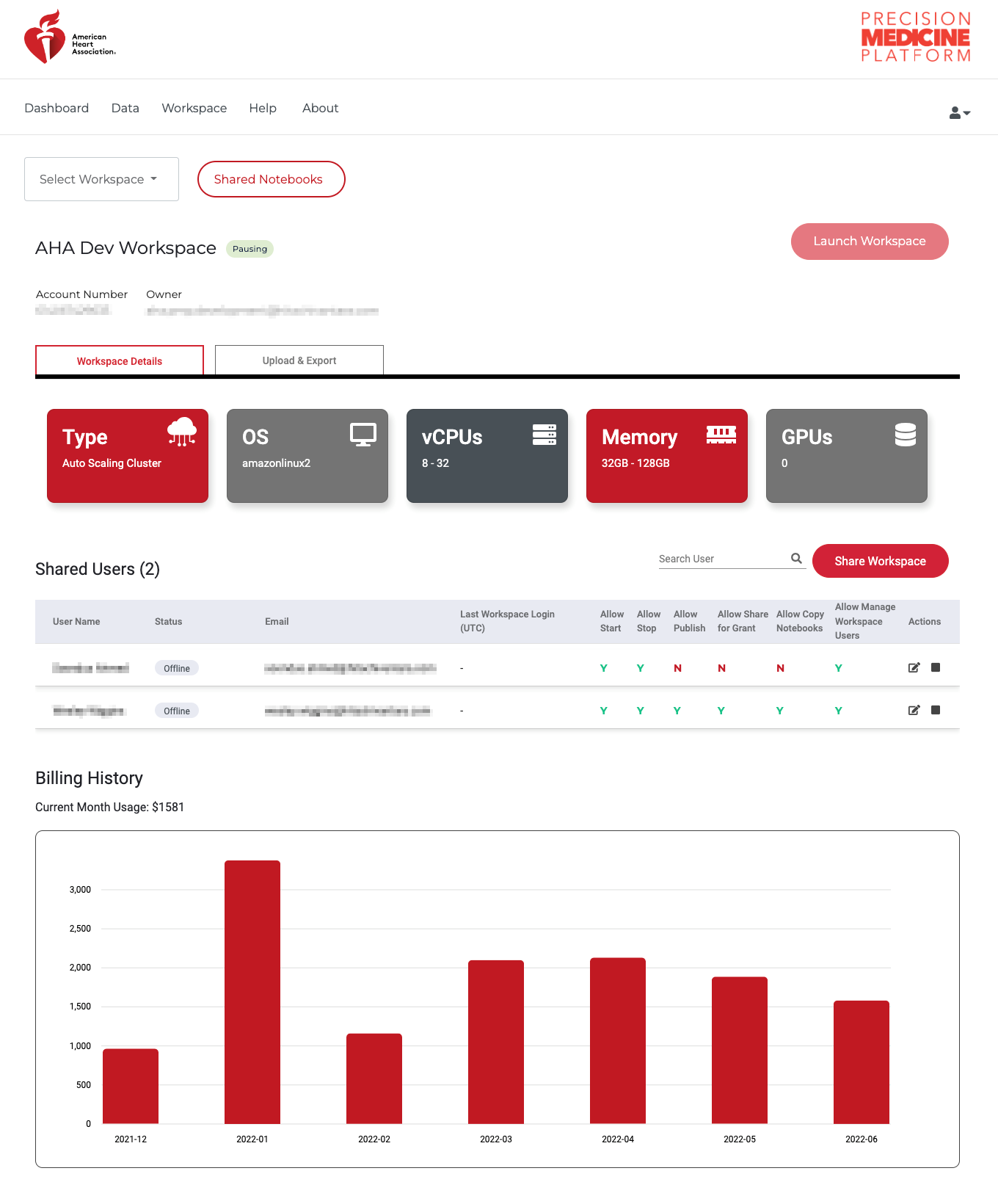
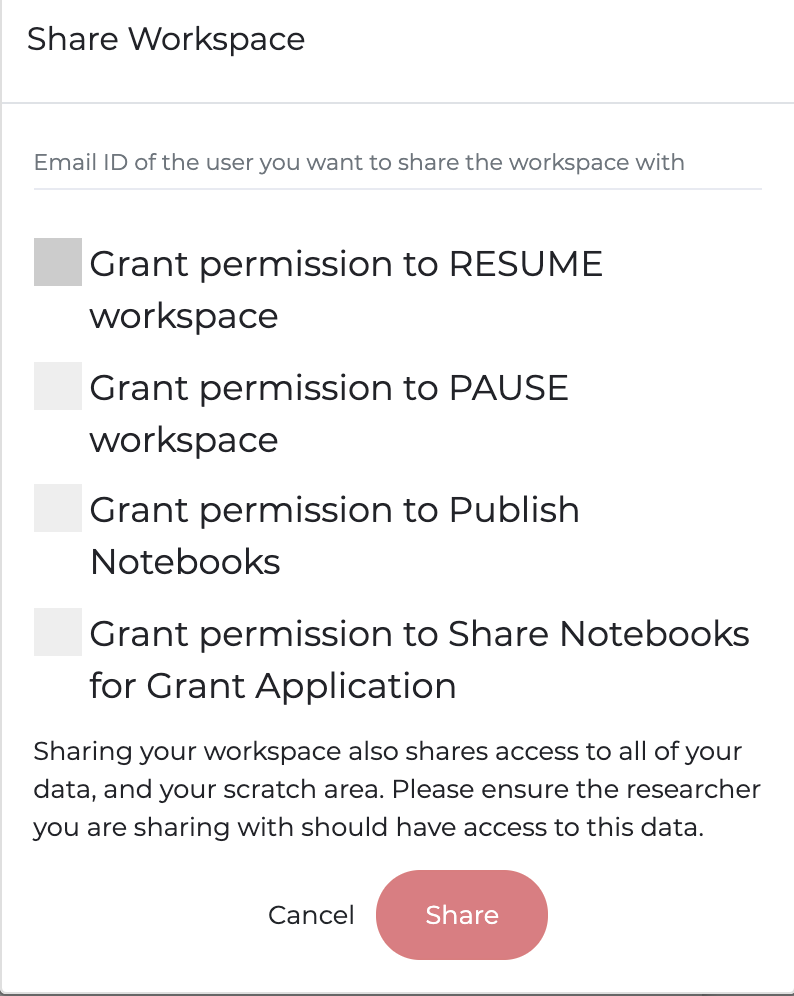
The design of PMP workspaces is such that when a user is provisioned a workspace, no one except the user has access (including the Association). All code and data is housed behind a secure, walled garden that cannot be accessed by anyone by the user. To facilitate collaboration and provide workspace access for teams working on projects, the owner of a workspace can share their workspace with other people who have a PMP account. Shared users can access all code and files in the workspace, and it is a workspace owners responsibility to ensure the user has the appropriate data approvals prior to sharing the workspace. To share a workspace with a user, click the menu icon on the workspace portal. Enter the email for the shared user on the PMP and set any desired restraints (e.g. allowing to share/publish code or allowing to start/stop the workspace)

The Association recommends pausing your workspace when it will not be in use for a few days or longer. This preserves the overall resources needed to run and maintain a workspace and allows for updates to be completed. A workspace can be paused by clicking the red pause button on the workspace portal. Additionally, users can change architectures and compute options when pausing and restarting. Upon restart the user will be provided with the option to configure and EMR, EC2-GPU, or EC2-CPU workspace. For custom sizing options, please contact us.
Pre-Installed Software
PMP workspaces come with R, Python, Scala, Spark, and are available as well as variety of standard packages and software used in clinical and biomedical analyses pre-installed. These include a variety of machine learning and statistical modeling packages and software such as Keras, Tensorflow, pytorch, and caret. Common genetic packages available through bioconductor and other command-line tools are also provided. A full list of packages and software available on the platform can be found here. The workspace leverages, Jupyter, JupyterLab and RStudio to provide a UI based environment for users to conduct analyses. The ReadMe notebook on the Jupyter Home page (/mnt/workspace/) provides information regarding workspace usage, including trouble shooting instructions, software references, and tutorials.
Customizing the workspace and installing Software
The workspace is also fully customizable. License softwares, such as SAS and MATLAB can be installed for users with a license. Python and R based packages can easily be installed by users via pip and install.packages() respectively. The workspace has a Red Hat, Linux Operating System. Yum can be used to get, install, and manage software packages. Users are granted root access to enable users to customize their workspace to fit their needs.
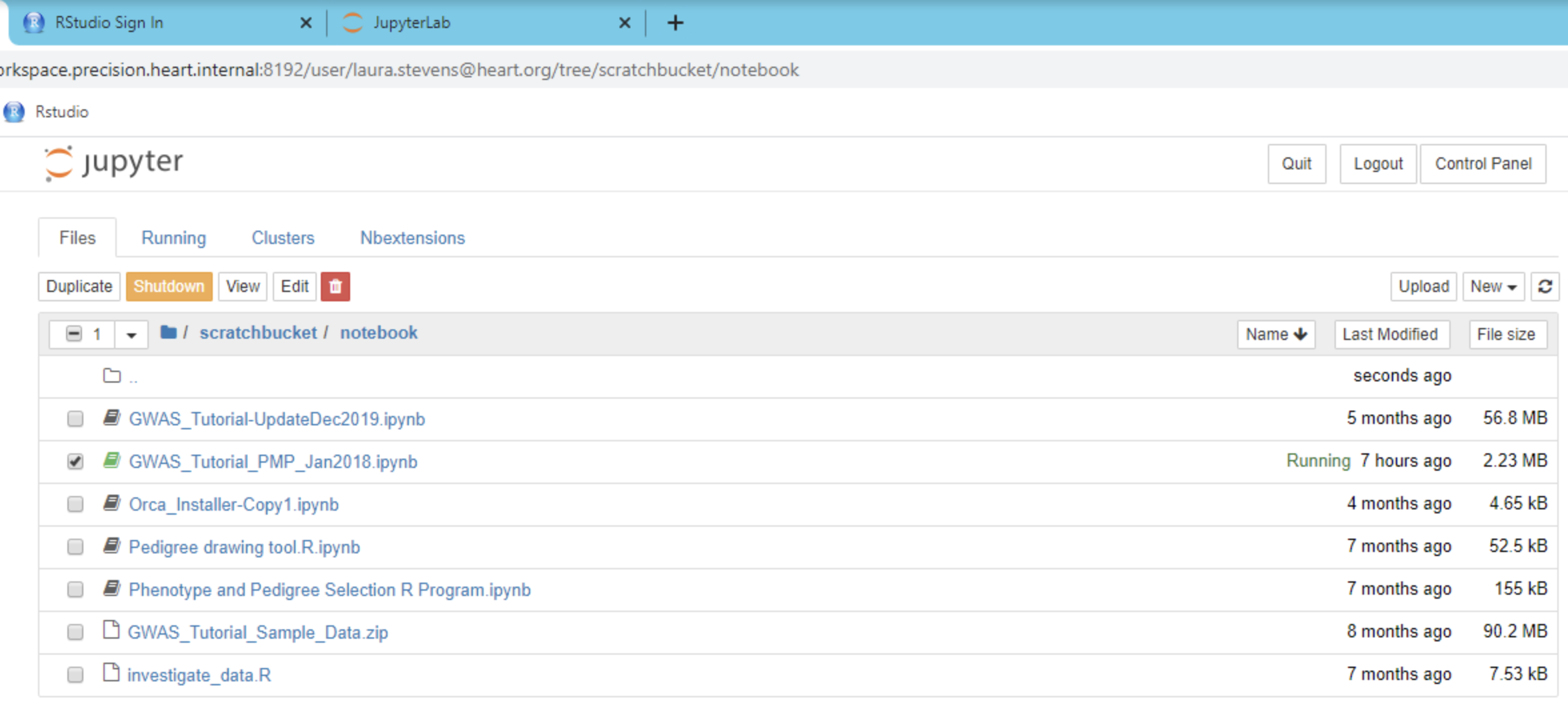
A variety of tutorials and examples are available on the Platform. Tutorials range from machine learning with Spark, to GWAS analyses to tutorials explaining software installation and fundamental computational concepts. The design of PMP workspaces is such that when a user is provisioned a workspace, no one except the user has access (including the AHA). All code and data is housed behind a secure, walled garden that cannot be accessed by anyone by the user. An overview of many of these tutorials is provided in the ReadMe notebook, additional tutorials and examples can be found in the jupyter_notebooks folder on the Home (/mnt/workspace) directory of the workspace. Users can also request data science and support services for help with informatic and data science analyses. For more information contact us.