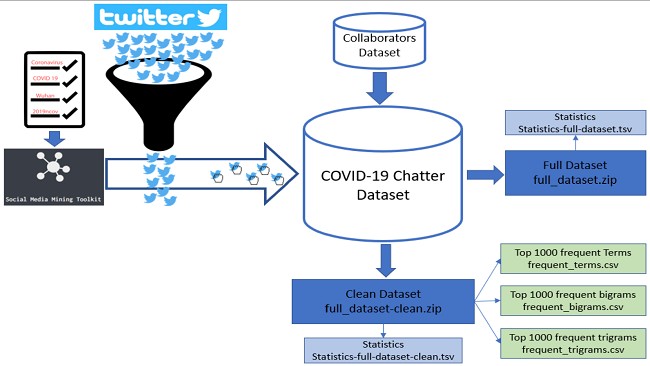
As the COVID-19 pandemic continues to spread, there is a need to facilitate data sources that measure the role of social dynamics of such a unique worldwide event for incorporation into biomedical, biological, and epidemiological analyses. For this purpose, we release a large-scale curated dataset of over 1.34 billion tweets, growing daily, related to COVID-19 chatter captured from 1 January 2020 to this day. We provide a freely available data source for researchers who have conducted more than 200 research projects as diverse as epidemiological analyses, emotional and mental responses to social distancing measures, the identification of sources of misinformation, among many others.
Lead/Mantainer: Juan M. Banda
Collaborators/Data providers: Gerardo Chowell, Ramya Tekumalla, Guanyu Wang, Jingyuan Yu, Tuo Liu, Yuning Ding, Katya Artemova and Elena Tutubalina.
The effort was launched during a conversation between Dr. Chowell (Epidemiologist) early January 2020 about capturing data for COVID-19. Ramya Tekumalla helped with a few initial scripts for pre-processing, and Guanyu Wang, Jingyuan Yu, Tuo Liu, Yuning Ding provided some earlier data we did nott have and additionally Katya Artemova and Elena Tutubalina provided data in Russian that we did not capture.
After version 5 of the dataset, Juan M. Banda took over the data management process creating automated collecting scripts, pre-processing scripts, and dataset release preparation scripts, which mostly automated the process. We release one new version of the dataset every week (hence the 122 versions so far!). While mostly an automated process, Juan M. Banda still has to initiate the process every weekend.
Over what period of time did this body of work occur and what were the goals of your project?
The project was planned to continue until the pandemic was over, so we are still making new weekly releases. Our original goals were to provide researchers with little experience using social media data with a curated dataset that they could pre-filter before hydrating (downloading the full tweets from Twitter). The hardest parts of collecting longitudinal datasets from Twitter involves ‘listening’ to the free Twitter stream 24 hours a day, seven days a week, and storing all acquired tweets on a local computer/server. By doing this for other researchers, they can focus on just ‘hydrating’ the tweets they want. We provide filtering fields for language, from the time ranges they want. Additionally, we released tools in the past to process tweets that are easier to use for non-computer scientists (https://github.com/thepanacealab/SMMT), and we provided tutorial code for any users https://github.com/thepanacealab/covid19_twitter/blob/master/COVID_19_dataset_Tutorial.ipynb.
What data sharing and/or reuse practices has your team adopted?
We shared everything in a Zenodo repository (https://doi.org/10.5281/zenodo.3723939) which handles versioning for each weekly release. Zenodo follows the FAIR principles for data sharing, and we only need to adhere to Twitter’s data sharing policies of not sharing full tweets.
What data sharing or reuse practices would you recommend all researchers adopt, and why?
In an ideal world, most resources would be shared using the FAIR principles, so they are easy to access, interoperable and can be replicated, and also use public repositories like Zenodo and Github, instead of putting data objects on personal websites or University websites.
What do you think is compelling about how you shared or reused data?
Most researchers share data only after they publish and extract as much as they can out of the data. We released data before we even had a clear research question to answer, this is a very different approach to most people. We wanted others to have access to this data to answer many questions and advance science instead of hoarding it to answer just one question. And this worked!
What existing standards and processes did you leverage in your approach?
We defined our own sharing standards around the terms and conditions of data sharing from Twitter. So while we can not share the full tweets text and other meta-data fields, we share enough for people to be able to filter tweets (tweets or retweets, and language) and avoid downloading unnecessary tweets which allows users to get to relevant tweets directly. We always release the same elements, and everything is fully standardized so each weekly release is exactly the same.
How could this approach be replicated by others?
All our code is publicly available at https://github.com/thepanacealab/covid19_twitter for anybody to review. We have over 175 forks from the code repository and it has been stared over 428 times.
I would tell them to do it as long as they follow the underlying data sharing rules. For us, releasing data before we even published anything opened a new world of collaborations and directions for our work. It has impacted countless other researchers, with nearly 180,000 downloads in Zenodo. We have published papers with groups from all over the world, like MIT (https://tinyurl.com/39z6h289), Canada (https://tinyurl.com/bdffmjkw), and published and worked with talented undergrads from the Harvard Coronavirus Visualization Team (https://doi.org/10.36190/2022.81). We used Twitter data to enhance Epidemiological models - one of our initial goals - (https://doi.org/10.1371/journal.pntd.0010228, https://doi.org/10.1186/s12879-021-06122-7). However, our proudest achievement is that over 200 papers (https://tinyurl.com/3zwerryb) that we are not part of have cited our work in a span of two years, and have worked with social science summer schools (BAY-SICSS - https://tinyurl.com/2s4s3sa3). One last important thing to mention is that our work has had all this impact, and nobody has funded us - we applied for NSF Rapid grants, Fast Grants, and other solicitations. We used our own time and resources, and continue releasing new versions.