
High-throughput molecular profiling technologies have revolutionized biomedical research. However, they also present significant challenges in analyzing omics data, especially for those with no/limited bioinformatics and statistical expertise. We hypothesize that common omics data analyses can be conducted effectively using natural languages. To test this hypothesis, our team has developed DrBioRight, an open-access, natural language-oriented analytics platform. This platform allows users to ask biological questions directly through natural language. It automatically understands users’ intentions, identifies the related datasets, performs diverse bioinformatics analyses, and returns the results in a timely, visually attractive way.
Our team has broad multidisciplinary expertise in cancer biology, bioinformatics, biostatistics, software engineering, and scientific training. Dr. Liang is a recognized leader in computational cancer genomics. Collaborating with Dr. Li, his team has maintained an outstanding track record in developing widely used bioinformatics tools, including The Cancer Proteome Atlas (>40,000 users), TANRIC (>27,000 users), MCLP (>15,000 users), SurvNet (>6,000 users), and FASMIC (>4,000 users). The team has worked together for the last 5 years on the three functional components of DrBioRight: (1) software development, led by Dr. Li, including model construction and training of natural language processing (NLP) modules, setting-up cloud computing architecture, and maintaining databases; (2) data analysis and interpretation, led by Dr. Liang, for developing analytic modules, exploring high-dimensional data, and identifying biological insights through data reuse; and (3) data management and software dissemination, co-led by Dr. Li and Dr. Liang, to collect, harmonize, and share the resources.
Development period and goals
We initiated our project in 2016 and released the first version in 2018. Our goals are to: (1) greatly increase research efficiency by minimizing the informatic barriers researchers face in mining complex cancer omics data; (2) substantially improve the quality and interpretability of omics data analysis; (3) vertically enhance the transparency and reproducibility of data processing and computational analysis; and (4) actively serve as an open-access, open-development hub to promote the integration and dissemination of widely used analytic tools and bioinformatics pipelines.
Our practices on data reuse
(1) We integrate natural language processing libraries and constantly improve AI models to build our query module, which identifies user intents and translates them into an executable bioinformatics analysis task. This feature reduces the data analysis barrier to a minimal level. (2) Our platform employs a chat interface, through which one can start a one-on-one conversation with the AI agent but also can invite collaborators to join a “group discussion” and explore the results together. (3) We integrate workflow and markdown languages to generate analysis reports, thereby ensuring that the executed analyses are transparent and the obtained results are reproducible. (4) We incorporate Docker and GitHub to build an open-access, open-development center that allows software dissemination and contributions to/from other bioinformatics researchers and software developers.
Our recommendation
There are two major types of bioinformatics tools for omics data exploration (1) “module hubs” provide graphics interface to assemble bioinformatics pipelines and perform user-defined tasks; and (2) “interactive data portals” focuses on easy analysis and visualization for preloaded datasets. A new paradigm for data sharing and reuse should interconnect module hubs and interactive data portals through a new class of “intelligent analytics,” powered by a natural language-based interactive interface.
The compelling part about our platform
The most compelling part is the natural language-oriented human-machine interactive interface. Unlike other analytic platforms with predefined algorithms and rules, DrBioRight uses user feedback as input to improve itself through the underlying AI models. This effort will automatically increase the accuracy of translating a user’s intents to appropriate analytic tasks and informative visualization.
Existing standards and processes used in our approach
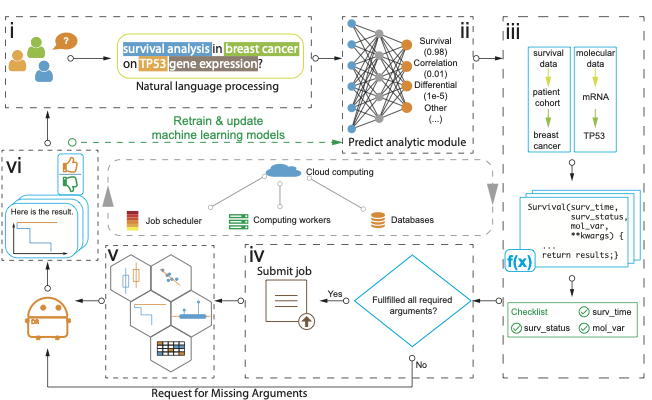
Our platform consists of two subsystems: a web interface and a backend compute server. Through a chat interface, users can simply type an omics data analysis question in the input area. The program will run its NLP module to tag the recognized entities (Fig. i). The backend AI module will predict the best-matched analytic task (Fig. ii). The program will then call the analytic module, identify the related dataset, and check if all required parameters are filled (Fig. iii). It will confirm with the user if the task is the user’s intended analysis. If confirmed, a job scheduler will submit it to a job queue and process the job (Fig. iv). Once the job is finished, DrBioRight will call a corresponding visualization module (Fig. v). Finally, the results will be sent to the user through the dialog area with a rating feature for each job (Fig. vi). The feedback will then be used as the training data to improve the performance of the NLP and the AI models.
Replicability
(1) We document our approaches in our previous publication and our official website, including written documentation and video tutorials; (2) we build an online system for submitting inquiries, nominating modules/datasets, and reporting bugs; (3) we develop web APIs to facilitate the integration of other widely used bioinformatics resources; and (4) we incorporate Docker and Gitlab to enable co-development and software disseminations with the community users.
One of our major goals is to increase the use of DrBioRight, promote interactions with its users, and facilitate the integration of data and other bioinformatics resources and tools. Specifically, we (1) create a repository of standardized pipelines for omics data; (2) build a developer center to support module contributions and dissemination; (3) provide written documentation, video tutorials, and hands-on workshops; (4) provide an online system for submitting software inquiries, nominating modules/datasets, and reporting bugs; and (5) develop web APIs and facilitate the integration of other widely used bioinformatics resources.
Promoting community engagement and outreach can substantially increase the user body and make a long-standing, positive impact, including (1) improving the AI and analytics platform based on user feedback; (2) enlarging the library of analytic/visualization modules by co-development; (3) compiling and harmonizing public omics datasets by providing public APIs; and (4) promoting research collaborations, thereby better serving the broader biomedical research community.