MSK-IMPACT is a targeted sequencing assay that has been used on >70,000 patients at Memorial Sloan Kettering Cancer Center (MSK) to identify genomic alterations in tumor samples, with the goal of providing diagnostic and prognostic insights, as well as identifying therapeutic targets to guide cancer treatment. All genomic and clinical data are shared within the institution and with the broader scientific community via the cBioPortal for Cancer Genomics, a web-based tool for data analysis and visualization. The MSK-IMPACT data have been used in 576 publications from MSK researchers alone, and these publications have been cited a total of 43,714 times, showing the impact this data sharing initiative has on the field of cancer research.
The MSK-IMPACT initiative is supported by a diverse team of researchers and clinicians with a shared commitment to sharing and disseminating data and knowledge. It was initiated with the establishment of the Center for Molecular Oncology (CMO) in 2014, led by Dr. David Solit, with the goal of sequencing a large number of tumor samples from a diverse set of cancer types, to enable discoveries that will ultimately lead to advances in precision oncology.
Michael Berger, PhD, an Attending in the Department of Pathology, developed the MSK-IMPACT assay. He and his colleagues, Dr. Marc Ladanyi, Dr. Rose Brannon, and Aijaz Syed lead a large team of software engineers and molecular pathologists to support the generation of sequencing results and to sign out molecular reports. Nikolaus Schultz, PhD, an Attending in the Computational Oncology Service, led the development of the cBioPortal, and his team is responsible for building and maintaining data pipelines, hosting the cBioPortal software, and developing new analysis and visualization features. With critical contributions from Benjamin Gross and Ino de Bruijn, the cBioPortal ingests MSK-IMPACT results on a daily basis and disseminates these results in real time to the MSK community.
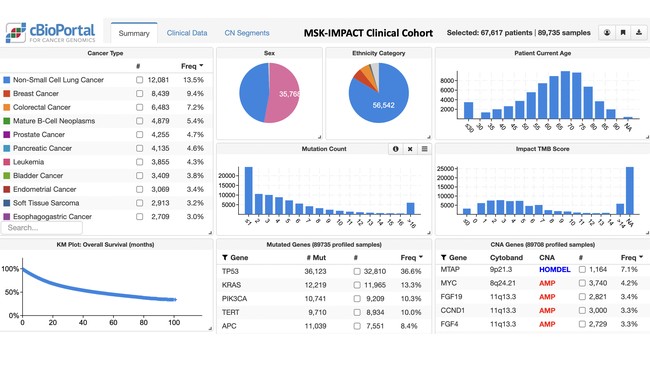
The use of the MSK-IMPACT assay for clinical use started in 2014 and is still continuing today. The MSK-IMPACT is a targeted sequencing assay that, by sequencing tumor and normal samples, can identify somatic and germline mutations in 505 cancer genes, as well as copy-number alterations and select gene rearrangements. The size of the solid tumor assay has grown from 341 to 505 genes over the years, and the heme-specific version of the assay now includes 468 genes. The data can be visualized and analyzed through the cBioPortal for Cancer Genomics, an open source software tool originally developed at MSK, which allows intuitive exploratory analysis of the molecular and clinical data.
To maximize the use of the MSK-IMPACT data, we chose to share the data via three main mechanisms:
1) All MSK researchers have access to the data through an internal instance of cBioPortal. The data set now comprises 89,735 tumor samples from 67,617 patients, and new data are added daily, at a rate of >12,000 samples per year. When researchers plan to submit findings based on the MSK-IMPACT data, they notify an internal publication committee, which reviews the abstract as well as proposed authorship to ensure proper credit for all contributors.
2) As manuscripts are published, we share the molecular and clinical data through the public instance of the cBioPortal, often along with rich clinical data. Data can also be downloaded via the cBioPortal Data Hub. These data can be used to validate findings or to make novel discoveries. To date, there are 576 publications based on MSK-IMPACT data from MSK researchers alone, and these publications have been cited a total of 43,714 times.
3) With a delay of a year, genomic data and specific clinical data elements from tumors sequenced using the MSK-IMPACT assay are shared via AACR Project GENIE, a multi-institutional data sharing project. Data can be accessed through a GENIE-specific instance of cBioPortal and through the Synapse platform.
The following data sharing practices have been adopted by our team:
We leveraged many of the data standards developed as part of The Cancer Genome Atlas (TCGA), a hugely successful molecular profiling study funded by the National Cancer Institute, which started in 2008 and ran for about a decade and generated detailed molecular profiling data from 10,000 tumor samples. These include data standards for somatic and germline mutations (MAF), as well as gene-specific and segmented DNA copy-number changes.
We also decided to use the cBioPortal for Cancer Genomics software, which was originally developed to support TCGA data, and we modified it to include capabilities for clinical data display.
Most importantly, we replicated the data sharing approach by TCGA, which made all genomic data available to the public immediately after generation. This was a key factor in the success of this program.
Since we used only open data standards and open source software, our approach can easily be replicated by others in the field. The cBioPortal software is already used by dozens of academic institutions across the globe, and our group is able to host data sets in the public instance of cBioPortal as different groups publish manuscripts based on their own tumor profiling data.
Data sharing has brought tremendous benefits to cancer research and patients. The impact of large sequencing projects such as TCGA cannot be overstated: they have improved our understanding of cancer, the way research is conducted, how the disease is treated in the clinic, and more.
AACR Project GENIE is another example of successful data sharing in the field. Established by MSK leadership, it is modeled after the MSK-IMPACT data sharing initiative and uses the same data standards, including our own tumor type ontology (OncoTree). MSK is the lead contributor with 40% of the data. GENIE also adopted the cBioPortal as its data sharing mechanism.
We have seen incredible benefits from having a shared resource at MSK, allowing clinicians and researchers to explore the data from all angles. Sharing a deep understanding of the dataset fosters collaboration across disciplines. Our effort has also improved the genomic literacy among clinicians: Because the cBioPortal was so powerful yet simple to use, clinicians started searching for their patients’ data among the deidentified institutional cohort, ultimately leading to the creation of a hyperlink to the cBioPortal from within each patient’s electronic medical record.