In the earliest days of the pandemic, dozens of labs across the world from the Human Cell Atlas initiative came together to understand what cells across the body are most vulnerable to the virus. Before it was possible to collect patient samples, we tapped into the Atlas, sharing both open and unpublished data, spanning 107 studies with more than 1.3 million individual human cell profiles. With new meta analysis we found which cells are the likely targets of the virus in the lung and airways, as well as in many other tissues. We related risk to age, sex, and smoking, found pathways that relate to emerging clinical phenomena, and laid the foundation to later build atlases of organs from patients who succumbed to COVID-19.
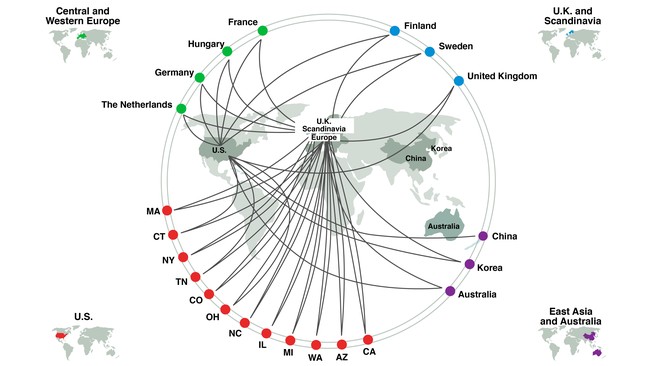
Our team had been building over the course of several years under the umbrella of the Human Cell Atlas (HCA) initiative, a grassroots, global, scientist-led project co-founded and co-led by Aviv Regev and Sarah Teichmann. Scientists interested in and involved in this initiative had already been meeting on a regular basis both at HCA meetings and separately, but in this project we rapidly coalesced around a shared goal, first focused on the lung and airways, and then expanding to the entire body. We collaborated at every level - graduate students, researchers, postdoctoral fellows and PIs. Our lung network and the researchers involved in this project spanned four continents, 11 countries, and 12 US states. Leveraging our established data sharing policy, governance and Infrastructure from the Human Cell Atlas, and deep personal trust in our network, a sub-team in the US and Germany managed all data, as well as established the analytical approaches needed for meta-analysis.
We began this work in early February 2020 and completed the analysis by April 2020, when we submitted our initial manuscript and posted it to bioRxiv, and released a portal with all relevant data. Our goal was to leverage existing single-cell data sets, published and unpublished, collected by our community and others to shed light on the poorly understood biology of SARS-CoV-2 infection at the time — what cells are likely to become infected, what other genes they express that could help us understand viral replication and the impact of SARS-CoV-2 on the body at its basic cellular level, and whether we could explain the vulnerability of particular segments of the population to infection and resulting illness. This effort relied on data collected across many grant initiatives in the US (including, NIH grants from HubMAP, LungMAP, GTEx, BRAIN, NCI Moonshot, and many individual NIH grants) and across the world. The Human Cell Atlas is committed to open data sharing and reuse, and many of our team members have long been proponents of open data and methods sharing, as well as posting of preprints prior to peer reviewed publication, all of which allow our field to move at a rapid pace in general, and particularly on this time-sensitive project. All members of the HCA agree to adhere to such principles of open sharing and collaboration, all HCA branded papers share their data, to the extent permitted by limits on data reuse or sharing and while maintaining patient or donor privacy. We have data sharing policies in place, a cloud-based Data Coordination Platform, and multiple open portals for querying, analysis and visualization by the entire community. The speed at which we completed this project is a testament to the benefits of not only open sharing and collaboration, but also to the value of collecting healthy reference datasets that can then be repurposed to understand disease. Moreover, this project formed a foundation of a parallel collaborative effort to profile, analyze and share data on samples collected at autopsies of patients who succumbed to COVID-19, another large-scale collaborative effort in the HCA community. By comparing to the samples from organs at autopsy to those of the healthy reference we made multiple discoveries on COVID-19 pathology.
We relied on existing standards in our community for sharing single-cell RNA-Seq datasets — including the data format, accompanying metadata, and sharing/uploading to publicly available data platforms, databases and portals. In our publication, we describe our methods to harmonize across datasets, which can serve as a basis for integrating multiple datasets that were collected separately to glean additional data from their power. These have now been expanded and extended as part of the HCA’s atlas integration efforts, for example in the Human Lung Cell Atlas. The work to optimally integrate across datasets within and across tissues is ongoing in the Human Cell Atlas community, and as these methods arise they will become easier to distribute widely to further enable this approach.
This project was a prime example of a new scientific question arising that we could not have imagined when our team members collected the data that originally sought to answer other questions, such as what cell types are in the lung, or what are the gene expression patterns across the airways and how might this relate to inherited diseases. We were able to build on the willingness of our community members to share the data they had collected — including some that had not yet been published — to answer pressing public health questions and help understand a new disease. This outcome demonstrates the value of exploring potential collaborations even before there is necessarily a concrete, timely project to pursue as a team, because these initial conversations can lead in unexpected directions.