Establishing consensus around the transcriptional interface between SARS-CoV-2 (SARS2) infection and human cellular signaling pathways can catalyze the development of novel anti-SARS2 therapeutics. Here, we used publicly archived transcriptomic and ChIP-Seq datasets to compute and validate consensus transcriptional regulatory networks of SARS2-infected cells. Our analysis accurately predicted aspects of the transcriptional impact of SARS2 infection that were subsequently published by much larger laboratories in high impact publications. Our consensus transcriptional regulatory networks are freely accessible through the Signaling Pathways Project knowledgebase, and as Cytoscape-style networks in the Network Data Exchange repository.
Dr Neil McKenna leads the team. Trained as a cell biologist, he has 20 years experience in data sharing and re-use initiatives. Reflecting this experience, he led the first data repository in the US to mint digital object identifiers as unique identifiers for datasets. He currently leads the Signaling Pathways Project (SPP), a trans-omics knowledgebase for cellular signaling pathways. His career goal is to give datasets parity of esteem with research articles.
Dr Jeff Grethe is PI of the NIDDK Information Network (DKNET) and a long time collaborator of Dr McKenna in various NIH and FAIR data initiatives such as BD2K and DataMed. His group maintains the cloud environment in which the T1D regulatory networks are hosted, as well as SPP.
Dr Scott Ochsner joined Dr McKenna's group in 2009 after graduate research in molecular reproductive biology. He's never met a dataset he didn't want to put through R and is happy to do the leg work required to throw a lens on to an RNA-Seq dataset to make sense of its biology.
Last but not least, Dr Rudi Pillich is a senior curator at NDEx, developed by the same group that brought you Cytoscape. Rudi makes all our letters and numbers look visually engaging and informative in the NDEx website
Period of time and goals
This work began when COVID lockdown enforced remote working at Baylor College of Medicine. The goal was to build consensus SARS2 transcriptional regulatory networks that accurately predicted the impact of SARS2 infection on human cell signaling pathway nodes. This goal was borne out when the results of our analysis were validated by large basic and clinical laboratories in high impact publications.
What data sharing and/or reuse practices has your team adopted?
The SARS2 consensus networks and their underlying data points and metadata, as well as original datasets, are freely accessible on the Signaling Pathways Project and NDEx web resources. Programmatic access to underlying data and metadata is supported by a RESTful API. All SPP datasets are biocurated versions of publically archived datasets, are formatted according to the FORCE11 Joint Declaration on Data Citation Principles, and are available under a Creative Commons CC BY 4.0 license. The original datasets are available are linked to from the corresponding SPP datasets using the original repository accession identifiers. Consistent with FAIR guidelines, SPP consensus networks are assigned digital object identifiers as persistent identifiers.
What data sharing or reuse practices would you recommend all researchers adopt, and why?
Annotate, annotate, annotate. Data are nothing without context. By all means ensure that your datasets are technically robust, but also provide as much information you can as around the biology of the dataset - genes or proteins involved, cell type, viral infection strains and more. All this information is tremendously valuable to curators.
What do you think is compelling about how you shared or reused data?
An effective research community response to the ongoing impact of SARS2 infection on human health demands routine access to computational analysis of existing datasets that is unhindered either by paywalls or by lack of the informatics training required to manipulate archived datasets in their unprocessed state. Moreover, the substantial logistical obstacles to high containment laboratory certification emphasize the need for fullest possible access to, and re-usability of, existing SARS2 infection datasets to focus and refine hypotheses prior to carrying out in vivo SARS2 infection experiments. We feel this work is a compelling example of how judicious use of existing datasets can catalyze research of direct relevance to human health.
To ensure that our efforts are broadly aligned with established community standards, we have adapted existing, mature classifications for receptors (International Union of Pharmacology, IUPHAR), enzymes (International Union of Biochemistry and Molecular Biology Enzyme Committee) and transcription factors (TFClass). On the technical level we make extensive use of open web technologies and application programming interfaces to ensure maximum interoperability with other resources. All our standard operating procedures have been extensively documented in the publications listed to facilitate replication by other resources. Consistent with emerging NIH mandates on rigor and reproducibility, we used the Research Resource Identifier (RRID) standard to identify key research resources of relevance to our study. Finally, we developed a YouTube tutorial “Visualization of the CoV transcriptional regulatory networks in the Signaling Pathways Project knowledgebase and Network Data Exchange repository” to assist researchers in the use of these resources to generate hypotheses around SARS2 infection of human cells.
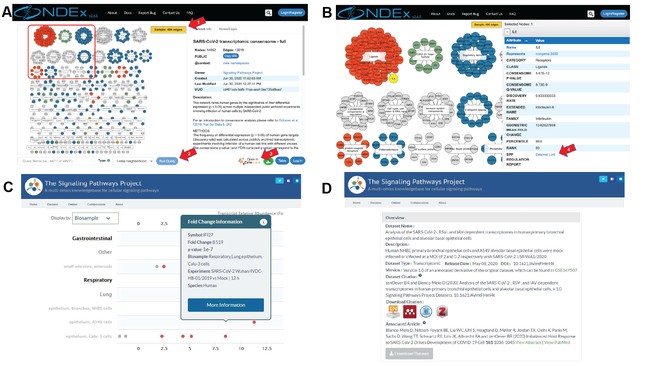
To maximize community engagement of our work, we made our networks freely available to the research community in the Network Data Exchange (NDEx) repository and the SPP knowledgebase. Individual networks can be browsed in NDEx in point and click Cytoscape-style interfaces (Image A&B). The integration in NDEx of the popular Cytoscape desktop application enables any network to be seamlessly be imported in Cytoscape for additional analysis. Target transcript symbols in NDEx networks link to SPP transcriptomic Regulation Reports in which clicking on a data point opens a Fold Change Information window (Image, C) that links to the SPP curated version of the original archived dataset (Image, D). Like all SPP datasets, SARS2 infection datasets are comprehensively aligned with FAIR data best practice and feature human-readable names and descriptions, a DOI, one-click addition to citation managers, and machine-readable downloadable data files. Reflecting the impact of our work on the research community, the resulting publication is in the Altmetric 98th %ile. We hope that our work is an inspiration to junior data scientists and has raised awareness among basic scientists as to the inherent and enduring value of dataset re-use.